|
In
the frame of the project “Phylogeny and survival strategies of photosynthetic
extremophilic microorganisms, PRIN-MIUR the aim of the research is to study
species of extremophilic terrestrial cyanobacteria adapted to conditions of
water stress and low irradiance. The first task is to characterize isolates of
the genus Chroococcidiopsis
isolated from hot and cold deserts and isolates of epilithic cyanobacteria from
hypogean environments, taking into account their molecular phylogeny and the
mechanisms underlying their ability to survive desiccation and low irradiance.
The biodiversity of these isolates is evaluated according two molecular
approaches : the first based on the analysis of the 16S rRNA sequence and of the
internal transcribed spacer (ITS). The second one is based on the employment of
a genomic PCR fingerprinting technique based on the use of primers derived from
cyanobacterial repeated sequences, namely short
tandemly repeated repetitive (STRR), long tandemly repeated repetitive (LTRR)
and the highly iterated palindrome (HIP1) sequences. This technique allows the
identification and typing of different isolates belonging to the same species as
well as provides evidence on the presence of repeated sequences in their genome,
giving thus information on its plasticity, contributing, thus, to highlight the
molecular mechanisms of their capability to survive to environmental stresses.
The second task is to evaluate the effects of various experimental conditions of
water stress and low irradiance by means of morphological characterizations and
analysis of the variation in their photosynthetic apparatus along with the
measurement of the photosynthetic activity in the various experimental
conditions. The third task consists in the characterization by HPLC and Circolar
Dicroism of the composition of the capsular polysaccharides (CPS) extracted from
the cyanobacterial cultures and in the evaluation of the polysaccharides
production and their biochemical characteristics in relation to water stress and
low-light availability. |
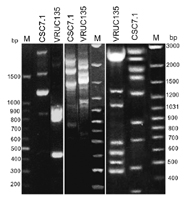
Genomic PCR fingerprinting of two Leptolyngbya strains with primers: (a) STRR2/STRR3, (b) STRR1/STRR3 and (c) LTRR1/LTRR2. The band profiles obtained allowed to distinguish between the two strains. Lanes M, 100 bp |